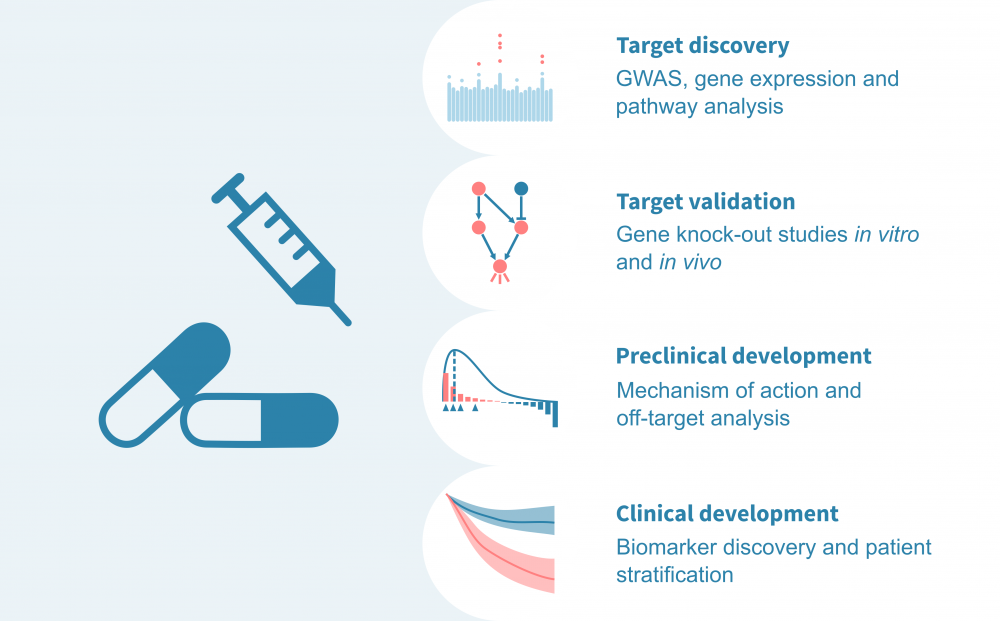
Discover drug targets, mechanisms and biomarkers with cutting-edge omics data analysis.
Molecular measurements based on next-generation sequencing (NGS) and mass-spectrometry (MS) are routinely used throughout the drug development process. RNA-sequencing and proteomics, in particular, reveal a more detailed view of pathways and functions in disease models and patients.
We work with customers developing small molecules and biologics as well as gene, cell and biomaterial therapies. Learn more about the stages in drug development that benefit from high-throughput molecular measurements coupled with state-of-the-art bioinformatics.